ELISA Competitive Inhibition Standard Curve
Competitive Inhibition ELISA Standard Curve
If you are using a non-competitive kit, please view our ELISA Standard Curve Guide.
Sandwich ELISA can be applied to the quantitative detection of target antigens or antibodies for antigens which present multiple epitopes. Small molecules or haptens do not present more than two binding sites for sandwich detection, and therefore sandwich method is not suitable for detection. In this case, competitive inhibition method can be used to detect the target molecules. The concept behind the competitive inhibition ELISA is that free HRP-labelled antigen will compete with the unlabelled antigen in the samples to bind the capture antibodies coated on the plate. The higher the sample antigen concentration, the weaker the signal. This is because the amount of labelled antigen bound to the plate is inversely proportional to the concentration of target molecules in the samples. After addition of the substrate solution, the intensity of color developed is inversely proportional to the concentration of target molecules in the samples. Small molecules such as hormones generally use this method.
Due to these differences, standard curve fitting is different for sandwich and competitive inhibition ELISAs. In the example below, the standard absorbance values for abx150385, Vitamin B12 ELISA Kit, are shown as a reference. As shown in Figure 1, the standard was diluted from 10000 pg/ml to 123.5 pg/ml. A negative control is also run. This contains no standard (0 pg/ml). For a competitive inhibition ELISA, the negative control well has the highest OD.
| Well | Concentration of standard (pg/ml) | Absorbance | Log of concentration |
| A | 0 | 3.747 | |
| B | 123.5 | 2.103 | 2.091666958 |
| C | 370.4 | 1.411 | 2.568670978 |
| D | 1111.1 | 0.878 | 3.045753148 |
| E | 3333.3 | 0.513 | 3.522874402 |
| F | 10000 | 0.159 | 4 |
Figure 1. Data of standard concentrations, absorbance values and Log of the standard concentration
For the competitive inhibition ELISA, there is no need to subtract the absorbance of the negative control. The O.D. value of the standard (X-axis) can be plotted against the log of the concentration of the standard (Y-axis), as an XY scatter plot, to get a 5-point curve. As shown in Figure 2, the X-axis is the Optical Density, and the Y-axis is the Log of the Standard Concentration.
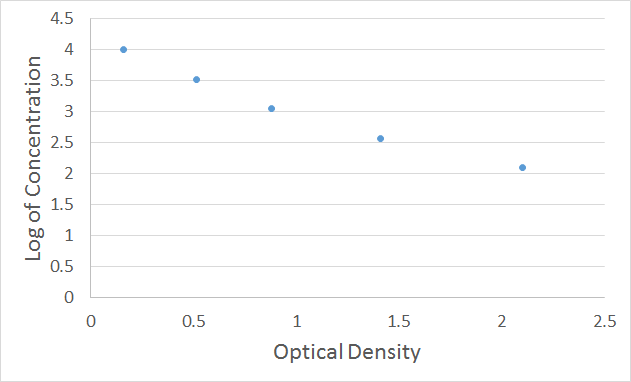
Figure 2. Plot of 7-point XY scatter
A polynomial order 2 trendline can be added to the graph. In Excel, this can be achieved by selecting any point in the XY scatter graph, right-clicking and selecting “Add Trendline”. The trendline formatting should then be amended to polynomial order 2 (the default trendline is linear) as shown in Figure 3 below. In the formatting section, further select “Display Equation on chart” and “Display R-squared value on chart”. Excel will then display the corresponding trendline formula and R-squared value on the graph. This is shown in Figure 4 below.
Figure 3. Addition and formatting of the trendline

Figure 4. Schematic diagram of a typical standard curve with trendline, equation and R-squared
In general, a good standard curve should have the following characteristics:
- R-squared value is greater than 0.95, and as close to 1 as possible.
- The OD of the highest standard of a competitive kit should be lower than 0.25.
- The maximum absorbance value should be higher than 0.8.
Troubleshooting
| Problem | Possible Source | Correction |
| Poor background | Contaminated negative control well | Change and use new pipette tips, containers and sealers |
| Poor R-squared | Improper standard curve preparation | Ensure accurate dilution procedure |
| Improper standard curve | Ensure accurate dilution of the standard | |
| Inaccurate Pipetting | Check and Calibrate pipettes | |
| Reused pipette tips, containers and sealers | Change and use new pipette tips, containers and sealers | |
| Low OD Values | Improper standard curve preparation | Ensure accurate operation of the dilution |
| Improper standard curve dilution | Ensure accurate dilution of the dilution | |
| Inaccurate Pipetting | Check and Calibrate pipettes | |
| Incorrect incubation times | Ensure sufficient incubation times | |
| Incorrect incubation temperature | Reagents balanced to room temperature | |
| Conjugate or substrate reagent failure | Mix conjugate and substrate; color should develop immediately | |
| No stop solution added | Follow the assay protocol in the kit manual |
